Sign up for CNN’s Wonder Theory science newsletter. Explore the universe with news about fascinating discoveries, scientific advancements and more.
The search for new antibiotics dates back to the Stone Age.
With the global population facing approximately 5 million deaths each year due to microbial resistance, according to the World Health Organization, the urgency to identify possible candidates has never been greater.
A research team led by bioengineering pioneer César de la Fuente is using artificial intelligence-based computational methods to extract genetic information from extinct human relatives such as Neanderthals and long-extinct ice age creatures such as the woolly mammoth and giant sloth.
Scientists say some of these small protein or peptide molecules they identified have bacteria-fighting powers that could inspire new drugs to fight infections in humans. The innovative work also opens up a completely new way of thinking about drug discovery.
“Their new sequences allowed us to uncover new types of molecules that we had not previously found in living organisms, expanding our thinking about molecular diversity,” said de la Fuente, assistant professor at the University of Pennsylvania. He heads the machine biology group. “Today’s bacteria have never encountered these molecules, so they may give us a better opportunity to target pathogens that are problematic today.”
The approach may seem like it’s out of left field, but experts say new ways of looking at the problem of antimicrobial resistance to existing drugs, a deadly and urgent problem for global health, are sorely needed.
“The world is facing an antibiotic resistance crisis. “My view is that a land, sea and air approach is needed to solve the problem, and if we have to go back to the past to provide potential solutions for the future, I’m all for it,” he said, majoring in molecular, cellular and developmental biology at the University of California, Santa Barbara. He did not participate.

Where antibiotics and alternatives might come from
Most antibiotics come from bacteria and fungi and were discovered by screening microorganisms living in soil. However, due to overuse in recent years, pathogens have become resistant to many of these drugs.
Scientists joining the global fight against superbugs are investigating different potential weapons, including phages, or viruses that nature created to eat bacteria.
Another exciting area of research involves antimicrobial peptides, or AMPs, which are infection-fighting molecules produced by many different organisms such as bacteria, fungi, plants, and animals, including humans. Mahan said AMPs have a wide range of antimicrobial properties against different pathogens such as viruses, bacteria, yeasts and fungi.
He added that while most traditional antibiotics work by focusing on a single target in the cell, antimicrobial peptides bind to the bacterial membrane in many places and disrupt it. According to Mahan, this is a more complex mechanism that could reduce drug resistance, but could also lead to increased toxicity due to the molecules’ potential to disrupt cell membranes.
There are a handful of peptide-based antibiotics in clinical use, such as colistin, which is made from a bacteria-based AMP. It’s a drug of last resort to treat certain bacterial infections because it can be toxic, Mahan said. A human AMP known as LL-37 has also shown potential.
Other promising AMPs have been found in unexpected places: pine needles and the blood of the Komodo dragon.
A ‘Jurassic Park’ moment
Over the past decade, De la Fuente has been using computational methods to evaluate the potential of a wide range of peptides as alternatives to antibiotics. The idea to look at extinct molecules arose during brainstorming in the laboratory when the blockbuster movie “Jurassic Park” was mentioned.
“The goal (in the movie) was to bring back the whole organisms, and obviously they had a lot of problems,” he said. His team began considering a more feasible idea: “Why not bring back molecules from the past?”
Advances in recovering ancient DNA from fossils mean that libraries of detailed genetic information about extinct human relatives and long-lost animals are now publicly available.
To find previously unknown peptides, the research team developed an artificial intelligence algorithm that recognizes fragmented regions in human proteins that may have antimicrobial activity. The scientists then applied this to publicly available protein sequences of modern humans (Homo sapiens), Neanderthals (Homo neanderthalensis), and Denisovans, another archaic human species closely related to Neanderthals.
The researchers then used the properties of previously described antimicrobial peptides to predict which of the newly identified ancient counterparts had the most potential to kill bacteria.
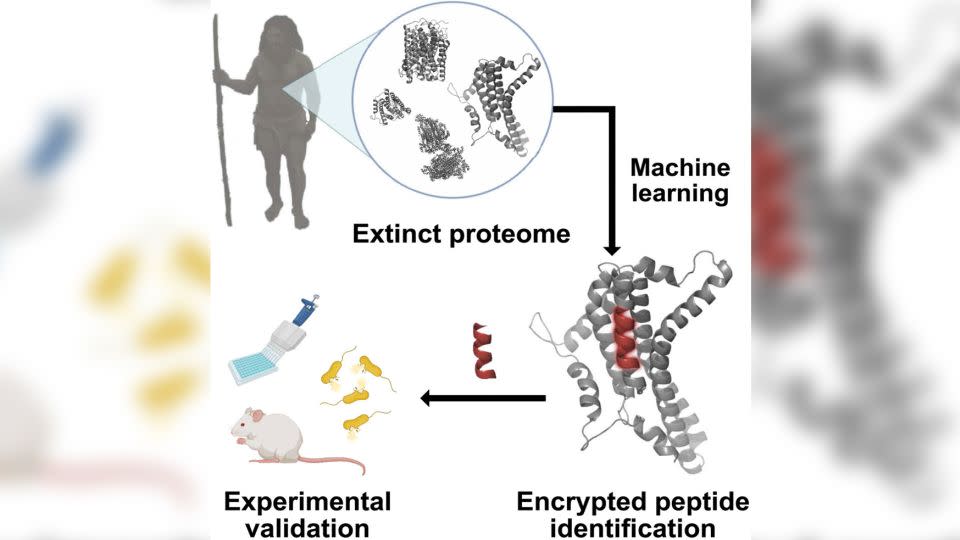

The researchers then synthesized and tested 69 of the most promising peptides to see if they could kill bacteria in petri dishes. The team selected the six strongest ones—four from Homo sapiens, one from Homo neanderthalensis, and one from Denisovans—and fed them to mice infected with the bacterium Acinetobacter baumannii, a common cause of hospital-acquired infections in humans.
“I think one of the most exciting moments was when we used chemistry to recreate molecules in the laboratory and then bring them back to life for the first time. From a scientific perspective, it was really cool to experience that moment,” de la Fuente said of the research, published in August in the scientific journal Cell Host & Microbe.
In infected mice that developed skin abscesses, the peptides actively killed bacteria; In those with thigh infections, the treatment was less effective but still stopped bacterial growth.
“The best (peptide) was what we call Neanderthal 1, which came from Neanderthals. That’s what was most effective in the mouse model,” de la Fuente said.
He warned that none of the peptides were “antibiotic ready” and would require a lot of modification. More important, he says, is the framework and tools his team developed to identify promising antimicrobial molecules from the past.
In research expected to be published next year, de la Fuente and his colleagues developed a new deep learning model to explore what he described as “extinction”—the protein sequences of 208 extinct organisms for which detailed genetic information was available.
The team found more than 11,000 previously unknown potential antimicrobial peptides unique to extinct organisms and samples from the Siberian woolly mammoth, the Steller sea cow (a marine mammal extirpated by hunting in the Arctic in the 18th century), the Steller sea cow, a 10-foot-tall species. promising candidates synthesized. long (3 meters) Darwin’s ground sloth (Mylodon darwinii) and the giant Irish elk (Megaloceros giganteus). He said the peptides they discovered exhibited “excellent anti-infective activity” in mice.
“Molecular extinction offers a unique opportunity to combat antibiotic resistance by resurrecting and harnessing the power of molecules from the past,” he said.
A strange but valuable approach
Dr., a group leader working on peptide antibiotics at the John Innes Center in the UK. Dmitry Ghilarov said the real bottleneck in the search for new antibiotics is not necessarily a lack of promising compounds, but getting pharmaceutical companies to develop them and test them clinically. Potential peptide antibiotics that may be unstable and difficult to synthesize. He did not participate in the research.
“I don’t see any pressing reason to look at paleoproteomes. We already have many of these peptides,” he said. “What we really need, in my view, is a deep understanding of the basic principles: what makes the peptide bioactive so we can design them.”
“There are many of these peptide antibiotics that are not developed and pursued by the industry due to challenges such as toxicity,” Ghilarov said.
Of the 10,000 promising compounds identified by researchers, only one or two antibiotic drugs have reached U.S. Food and Drug Administration approval, according to a paper published in May 2021.
D., a professor and associate director of research at George Mason University’s School of Systems Biology in Fairfax, Virginia. Monique van Hoek said the idea of molecular extinction was “a really interesting approach.” She did not participate in either study.
It’s rare for a peptide found in nature — whether extinct or from a living organism — to directly lead to a new type of antibiotic or other drug, Van Hoek said. More often, he said, the discovery of a new peptide will offer a starting point for researchers, who can then use computational techniques to refine and optimize the peptide’s potential as a drug candidate.
Van Hoek’s research is currently focusing on a synthetic peptide inspired by a naturally occurring peptide in the American alligator. The peptide is currently undergoing preclinical testing.
“It’s going really well so far. It’s very exciting because a lot of the other peptides I’ve worked on over the years have failed for one reason or another,” he said.
It may seem odd to look to crocodiles or extinct humans for a new source of antibiotics, Van Hoek said, but the magnitude of the crisis makes the approach worthwhile.
De la Fuente agreed. “I think what we need is as many new and different approaches as possible, which will ultimately increase our chances of being successful,” he said.
“I think we can find a lot of potentially useful solutions by looking back.”
For more CNN news and newsletters, create an account at CNN.com